Pan JBrowse
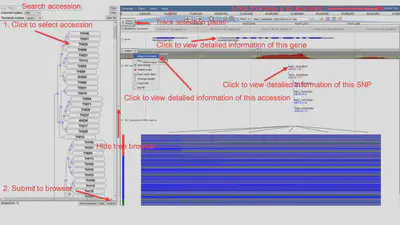
The Pan JBrowse page contains two parts, a dynamic tree browser on the left panel and a genome browser on the right panel. The tree was constructed from the SNP data. Users can select multiple nodes (including leaf nodes and internal nodes) and click the “Submit” button to visualize these rapeseed accessions in the genome browser. The tree browser also supports search function to accelerate target genome selection. The pan-genome reference sequence, gene annotation and TH001 sample’s presence frequency are three basic tracks. There are 2885 rapeseed genome tracks. Users can select any number of accessions data through the hidden “Select tracks” panel or the tree browser (only available for 381 high-depth accessions) as well. For the performance concern, we recommend to select less than 100 tracks each time.
The tree browser is composed of a tree viewer and 4 toolbars, one of which lies at the bottom of the browser. The top toolbar is for locating the terminal nodes by accession codes.
The next two toolbars change the behaviors of internal nodes and leaf nodes respectively. When "fold" is chosen, clicking internal nodes hides their child nodes. When "select" is chosen, clicking nodes select their child nodes (or themselves when clicking leaf nodes). A selected leaf node will be shown in genome browser when the "submit" button is clicked. When "preserve" is chosen, clicking nodes preserve their child nodes (or themselves when clicking leaf nodes). A preserved leaf node won't be hidden when folding its ancestors. Clicking a node for the second time behave oppositely in every chosen 'mode'.
The last toolbar lies at bottom. It provides functions on selection. Clicking the "Next Selected" button scrolls the tree browser down to the location of the next selected leaf node. Clicking the "clear" button deselects all selected accessions. Clicking the "submit" buttons shows selected accessions in genome browser. Clicking the "Help" button shows description about the usage of each button. Clicking the "Hide All" button hides all internal nodes.
The genome browser was based on JBrowse. The detailed usage of JBrowse could be acquired in the JBrowse official site.
There are two buttons on the top of this panel.
- Screenshot: generates a screenshot for JBrowse in PDF format.
- Share: share the link of current genome browser.
There are four types of tracks, including reference sequence, gene, accession coverage and SNP, and the first three types are default tracks.
- Reference sequence: the pan-genome sequence. Users could zoom in to see sequence at base level.
- Gene: the pan-genome gene annotation. Users could left-click to view detailed information.
- Accession Coverage: there are 2885 accession tracks.
- SNP: the 381 high-depth and 2504 low-depth SNPs.